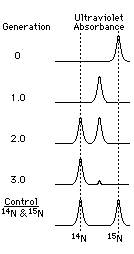
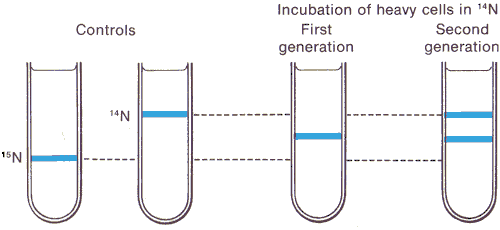
|
C.
Replication-
General Features |
|
|
1.
Bidirectional |
|
|
|
|
|
|
 |
|
|
|
a. Origin(s) of Replication- proceeds in both directions
|
|
|
|
b.
Replication
fork- where DNA helix splits into 2 ssDNA |
|
|
|
|
|
|
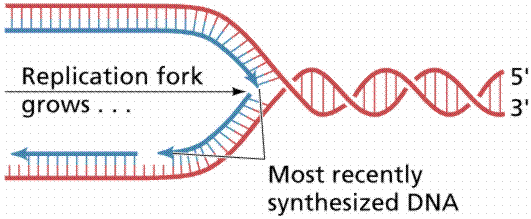
|
|
|
|
|
|
|
|
|
|
2.
Unwinding DNA |
|
|
|
a.
DNA
Gyrase- (Type II Topoisomerase)- introduces negatives coils with ATP |
|
|
|
|
-type I cuts
only ssDNA, type II cuts dsDNA
|
|
|
|
|
|
Structure
of DNA Gyrase,
Anitbiotic
resistance in topoisomerase,
DNA
Gyrase Inhibitors |
|
|
|
|
|
|
|
|
|
|
b.
Helicase-
Unwinds the DNA by breaking the hydrogen bonds |
|
|
|
|
-disrupts H-bonds instead of breaking phosphodiester
bonds (gyrase) |
|
|
|
|
-requires ssDNa
for binding (see replication process)
|
|
|
|
|
|
|
|
|
|
|
c.
SSB
(single stranded DNA-binding protein) bonds with single stranded to
prevent the double helix from recombining.
|
|
|
|
|
|
|
|
|
|
|
d.
Primase-
binds to open DNA and synthesizes an RNA primer |
|
|
|
|
- Once for leading strand, many times on lagging
strand
|
|
|
|
|
|
|
|
|
|
|
e.
DNA
polymerase- builds the "daughter strand" from the parent strand template |
|
|
|
|
-builds only in
the 5' to 3' direction |
|
|
|
|
-needs to have the 3' OH exposed to add new
nucleotide bases |
|
|
|
|
1.
Leading strand- parent strand that is 3' -->
5' direction, copied continuously |
|
|
|
|
2.
Lagging
strand- copied in a discontinuous mode, short segments |
|
|
|
|
|
-Okazaki fragments- ~1000 to 2000 nucleotides in
length
|
|
|
|
DNA Polymerase Tutorial |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
f.
DNA
Ligase |
|
|
|
|
-responsible for joining Okazaki fragments together
on the lagging strand |
|
|
|
|
|
|
|
|
D.
DNA
Polymerase
|
|
|
1. Properties
|
|
|
|
a.
selects
the appropriate nucleotide base that complements template bases
|
|
|
|
b.
builds
in the 5' --> 3'
direction and is antiparallel to the template
|
|
|
|
c.
cannot
initiate DNA synthesis- requires an oligonucleotide prime with free 3'-OH
|
|
|
|
|
|
|
|
|
|
2.
Types & Function
|
|
|
|
a.
DNA Polymerase I- (from E.coli)
|
|
|
|
|
1.
Function as polymerase
|
|
|
|
|
|
- able to catalyze DNA synthesis- in vitro- if
supplied with deoxynucleotide-5'-triphosphates (dATP, dTTP, dGTP, dCTP),
a template DNA strand and a primer
|
|
|
|
|
|
-
pyrophosphate is released when a new nucleotide base is added to a
pre-existing nucleotide
|
|
|
|
|
|
-
falls off about every 20 bases = poor processivity
|
|
|
|
|
|
|
|
|
|
|
2.
Function as exonuclease
|
|
|
|
|
|
-serves as a proof-reader and editor of 3' and 5' ends
|
|
|
|
|
|
|
|
|
|
|
b.
DNA
Polymerase III
|
|
|
|
|
|
-functions
as part of a larger complex- DNA Polymerase III Holoenzyme
|
|
|
|
|
|
-contains
10 subunits that function to increase the processivity- ~4.6 Mbase
|
|
|
|
|
|
-g
subunit- clamp loader
|
|
|
|
|
|
-b
subunit- sliding clamp
|
|
|
|
|
|
|
|
| R. A. Bambara, R. S.
Murante, and L. A. Henricksen (1997) Enzymes and reactions at the
eukaryotic DNA replication fork. J. Biol. Chem. 272:
4647-4650. Click
here for copy of paper. |
|
|
|
|
|
|
|
III.
Replication-
Procedure
(E.coli model)
|
|
|
Video of Replication
|
|
|
A.
Initiation
|
|
|
|
1.
Replication begins when DnaA protein attaches to oriC
(origin site)
|
|
|
|
|
-wraps
up the origin and opens the DNA
|
|
|
|
|
-the
opening of the 45 bp DNA is ATP-dependent
|
|
|
|
|
|
|
|
|
|
|
2. DnaB protein binds to the replication fork --> contains helicase and DNA gyrase
activity
|
|
|
|
|
|
|
|
|
|
|
3.
ssBP binds to ssDNA
|
|
|
|
|
|
|
|
|
|
|
4.
Primase synthesizes primers that are complement to DNA nucleotides
|
|
|
|
|
|
|
|
|
|
|
5.
DNA polymerase binds to both templates to begin DNA replication
|
|
|
|
|
|
|
|
|
|
B.
Elongation
|
|
|
|
1. DnaB advances the replication fork along in 2
directions
|
|
|
|
|
|
|
|
|
|
|
2. DNA Polymerase synthesizes new DNA strands along
the leading strand in the 5'-->3' direction.
|
|
|
|
|
-lagging
strand has to loop to maintain DNA Polymerase III Holoenzyme complex
|
|
|
|
|
|
|
|
|
|
|
3. Primase has to continually produce primers on the
lagging strand for each Okazaki fragment
|
|
|
|
|
|
|
|
|
|
|
4. DNA Polyermase I excises RNA primers, replaces
with DNA nucleotides and edits
|
|
|
|
|
|
|
|
|
|
|
5. DNA Ligase seals the nicks between Okazaki
fragments
|
|
|
|
|
|
|
|
|
|
C.
Termination
|
|
|
|
1. Ter
Locus (termination base sequence)
|
|
|
|
|
-also
requires a Tus protein (replication termination protein)- inhibits
the ATP-dependent DnaB helicase activity
|
|
|
|
|
|
|
|
|
|
|
2. Topoisomerase II (DNA gyrase) catalyzes the
excision of the new doubles helices which
splits them for each daughter cells produced during fission
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Additional Links: |
|
|
DNA
Replication & RNA Transcription |
|
|
DNA
Replication: Learning Objectives and Guiding Questions |
|
|
Chromosome
formation: coiling of DNA |
|
|
DNA Structure
and Replication |
|
|
Polymerase Chain
Reaction |
|
|
|
|
|
|
|
|
|
|
|
|