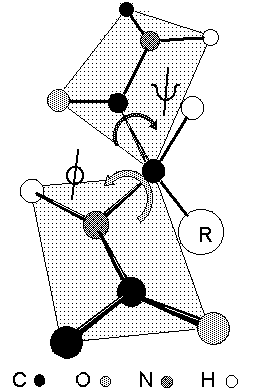
Rotation through the a-carbon

| Bond | Rotation | Torsion angle defined |
|---|---|---|
| free | phi | |
| free | psi | |
| C=O to NH (peptide bond) |
rigid planar due to double bond character |
omega |
Peptide bonds are almost invariably fixed at omega = 180o
or trans based on the relative alignment of C![]() atoms on either side of the peptide bond.
atoms on either side of the peptide bond.
http://employees.csbsju.edu/hjakubowski/classes/ch331/protstructure/phipsi.gif
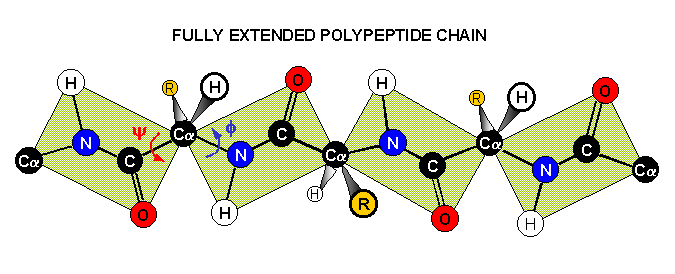
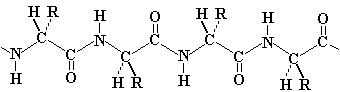
The rigidity of the peptide bond limits the number of arrangements that Pauling's models could fit without distorting bonds or forcing atoms closer than van der Waals radii would allow. Without this constraint, the peptide would be free to adopt so many structures that no single consistent pattern would emerge. By reducing the degrees of freedom, a well defined set of states emerges.
Pauling found that two general patterns conformed with atomic geometry:
1) an extended state for which angles phi = -135o and psi = +135o; the polypeptide chain alternates in direction, resulting in a zig-zag structure for the peptide chain. Note the shaded circle around R; the extended strand arrangement also allows the maximum space and freedom of movement for a side chain. The repeat between identically oriented R-groups is 7.0 Å, with 3.5 Å per amino acid, matching the fiber diffraction data for beta-keratins.

The helical form models could be built with varying degrees of twist, but one model fit the atomic dimensions especially well:
Alpha helix: has 3.6 amino acids per turn of the helix, which places the C=O group of amino acid #1 exactly in line with the H-N group of amino acid #5 (and C=O #2 with H-N #6).
