A.
Why Biochemistry?
Chemistry is the logic of Biological Phenomena……… Huh?
1.
Hierarchy of Life: Living
Organisms can be described by levels of complexity.
a.
Organism
b.
Systems- Respiratory, Circulatory, Lymphatic, etc.
c.
Organs- heart, lung, kidney, etc.
d.
Tissues-muscle, connective, epithelial, nerve
e.
Cells- prokaryotic, eukaryotic, specialized eukaryotic, RBC
f.
Organelles & Membranes
g.
Macromolecules
1.
Supramolecular complexes- Ribosomes, cytoskeleton, enzyme complexes
2.
Macromolecules- proteins, nucleic acids, polysaccharides, lipids
3.
Building blocks- amino acids, nucleotides, monosaccharides, fatty acids
4.
Metabolites- pyruvate, citrate, succinate, phosphoglyceraldehyde
5.
Inorganic Precursors- CO2, NH3, H2O, N2
h.
Atoms
2.
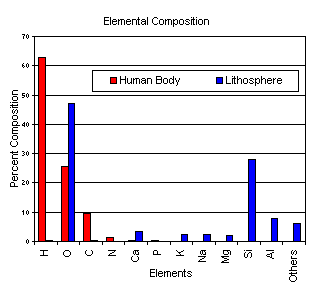
Composition of Biomolecules (In Human body by % composition)
a.
Hydrogen 63%
b.
Oxygen 25.5%
c.
Carbon 9.5%
d.
Nitrogen 1.4%

a.
Ability to form stable covalent bonds with itself- long chains
b.
Tetrahedral arrangement creates vast number of possible molecules
4.
What functional groups are prevalent in biomolecules?
a.
Amino
b.
Hydroxyl
c.
Carbonyl
d.
Carboxyl
e.
Amide
f.
Ester
g.
Phosphate ester
h.
Hemiacetal
i.
Acetal
B.
Proteins: An overview
The most prevalent group of biomolecules (50 % of dry body weight) and
the most diverse
1.
Types of Proteins
a.
Enzymes- Ribonuclease, Trypsin, Alcohol Dehydrogenase
b.
Regulatory- Insulin, lac Repressor
c.
Transport- Hemoglobin, Serum Albumin
d.
Storage- Ovalbumin, Casein, Ferritin
e. Contractile & Motile- Actin/Myosin, Tubulin/Dynein
f. Structural- a-Keratin,
Collagen, Elastin
g.
Scaffold- (used during cellular response mechanisms)
h.
Protective & Exploitive- Immunoglobulins, Thrombin/Fibrinogen, Ricin,
Venoms
i. Exotic- Monellin, Glue Proteins
2.
How many proteins are required for life?
a.
Mycoplasma genitalium (causes clamdyia)- 468 proteins
b.
Haemophilus influenza (causes bacteria meningitis)- 1703 proteins
c.
Escherichia Coli- ~4000 proteins
d.
Yeast- ~5000 proteins
e.
Worms- ~20,000 proteins
f.
Humans- 50,000-100,000 proteins
C.
Amino Acids
1.
General Structure-
a. a-carbon
(central carbon)
b.
a-Amino
Group
c. a-Carboxyl Group
d.
Hydrogen
e.
Side Chain (R)- Gives rise to variability in amino acids
The a-carbon,
a-Amino,
a-Carboxyl,
& Hydrogen are constant
for all
amino acids (except Proline)
| Typical Structure of an Amino Acid |
|
|
| R represents the side chain |
2.
Common Amino Acids
|
Common Amino Acids by Properties. Names, 3-Lettered Abbreviations, 1-Lettered Abbreviations |
|||
|
Nonpolar or Hydrophobic |
Polar, Uncharged |
Acidic |
Basic |
|
1. Alanine (Ala, A) |
1. Glycine (Gly, G) |
1. Asparatic Acid (Asp, D) |
1. Histidine (His, H) |
|
2. Valine (Val, V) |
2. Serine (Ser, S) |
2. Glutamic Acid (Glu, E) |
2. Lysine (Lys, K) |
|
3. Leucine (Leu, L) |
3. Asparagine (Asn, N) |
|
3. Arginine (Arg, R) |
|
4. Isoleucine (Ile, I) |
4. Glutamine (Gln, Q) |
|
|
|
5. Methionine (Met, M) |
5. Threonine (Thr, T) |
|
|
|
6. Proline (Pro, P) |
6. Tyrosine (Tyr, Y) |
|
|
|
7. Phenyalanine (Phe, F) |
7. Cysteine (Cys, C) |
|
|
|
8. Tryptophan (Trp, W) |
|
|
|
e. "21st" Naturally occurring amino acid: Selenocysteine. (Sec, U). See Selenocysteine.

Review: Biochemistry Quiz on Amino Acids
Resource: Properties of Amino Acids
3. Properties
a. Acid-Base Properties
-Amino Acids are polyprotic acids (release more than one H+) =
Dissociable protons
The
amino functions as a Lewis Base & carboxyl functions as a Lewis Acid,
therefore all amino acids undergo an internal Acid-Base Reaction where the H+ is
dissociated from the carboxyl forming a carboxylate and attached through a
coordinate covalent bond to the a-nitrogen
thus forming an ammonium ion.
-The
dipolar molecule formed (being electrically neutral) is called a Zwitterion
(German:
“Hybrid ion”).
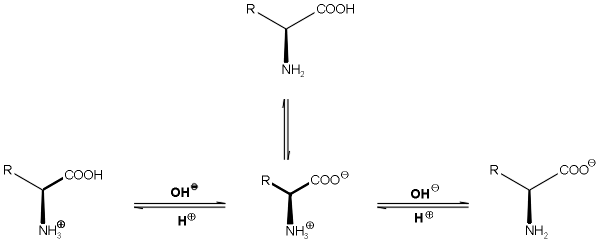
Cationic Form Zwitterion Anionic Form
Tutorial: Review of Acid & Base Theory
b. Stereochemistry
1. D,L- system for identify stereoisomerization
re: Chiral compounds are identified by the fact that they rotate the plane of polarized light
Three types of notation are used to describe chiral compounds:
a. (+) & (-) refer to the direction of rotation of polarized light.
b. R and S refer to the absolute (vs. relational) stereochemical relationship of the four substituents at a
chiral carbon. (Used by Organic Chemists)
c. D and L refer to the biochemical (chemical) relationship between glyceraldehyde (the simplest sugar
with an asymmetric center) and other compounds derived from it.
D- Dextrorotary- clockwise rotation of plane-polarized light
L- Levorotary- counterclockwise rotation of plane-polarized light
Fischer-Rosanoff Convention (or Rosanoff Convention): An arbitrary convention according to which
(+)-glyceraldehyde, now known to be (R)-2,3-dihydroxypropanal, was named D-glyceraldehyde (with the
enantiomer L-glyceraldehyde and its racemate D-glyceraldehyde) and taken to have the absolute configuration
represented by the Fischer projection formula shown below.
L-glyceraldehyde
Almost all amino acids that comprise proteins are in the L form.
replacing the D,L system due to amino acids with multiple chiral carbons
Study Aid: The R and S Convention for Absolute Configuration
All
a-carbon
configurations of the L-amino acids are (S) except for cysteine, by virtue of
the thiol group, and it is (R ).
3. Special Note. Fisher
Projection models
These are two dimensional drawing that represent 3-dimensional
structures.
|
|
Substituents above and below are behind the plane with a-carbon |
|
Substituents left and right are in front |
|
|
What enantiomeric form of alanine does this represent?
|
|
|
Can you draw the other enantiomeric form of alanine? |
|
1.
Primary
Structure- Linear sequence of amino acids (residues)
a.
Peptide bond- bond between carboxyl and adjacent amino groups. Formed through a condensation reaction
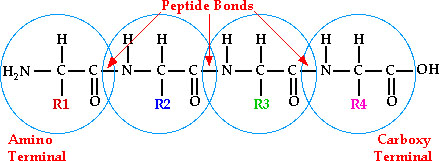
all primary sequences are read from the N-terminus to the C-terminus.
So
if R1 = methionine, R2= threonine, R3 = proline and R4 = valine, then the
sequence is named
met-thr-pro-val or can be named
MTPV
Characteristics of the peptide bond (amide linkage)
- coplanar-- no rotation (means some sort of pi bond electrons)
- polar
- resonance exists between C=O and C-N
A typical C-N bond is 1.43 pm and C=O is 1.23 pm. The diagram is showing that the C=O is
a little longer, and the C-N is a little shorter. This supports the resonance model.
- the phi/psi bond angles within a polypeptide allows for secondary structuring. Image.
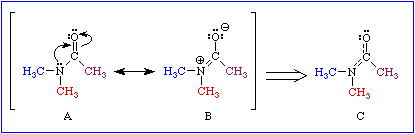
b.
Classification of peptides
1. dipeptide- 2 amino acid residues.
-named
from the N-terminal residue (add –yl instead of –ine) with
C-terminus residue being last
2.
tri, tetra, penta, etc. -peptides. (Same
game as dipeptides)
3.
Oligopeptides – usually contain 12-20 residues.
4. Polypeptides- consists of dozens of residues.
Resource: RCSB. Protein Data Bank. This is a search tool for finding protein structures knowing proteins
Resource: BLAST Searching from NCBI. This tool searches for proteins knowing sequences. Also can search for nucleic acids.
2. Secondary Structure- Regular patterns that occur due to interactions between the a-amino (-H) and a-carboxyl groups (-O). Image
Resource. Types of secondary structures.
a.
Alpha Helix-
coiled primary sequence found in many proteins
1. One turn of the a-helix represents 3.6 residues (13 atoms along primary
chain)
- Commonly referred as the 3.613
helix
2. Each residue extends 1.5 A along the
a-helix axis
-
amounts to 5.4 A per turn (3.6 residues x 1.5 A/residue)-
- this length defines the pitch
of the a-helix
3. The diameter of the a-helix is ~6 A
4.
Each peptide carbonyl is hydrogen bonded to the peptide (N—H) group four
5.
Each side chain is oriented from
the a-helix axis
6. Each peptide bond has a dipole moment, therefore with the uniform alignment of carboxyl oxygen, the helix itself
has a substantial dipole moment with a positive charge at the N-terminus and a negative charge at the C-terminus.
7. Other helical structures: image comparing helixes
a. 310 helix
b. 27 ribbon
c. 4.416 helix (p helix)
b. b-sheet
1. side-by-side peptide strands with H-bonds that form a “pleated sheet”
- the a-Carbons are found along the pleats of the sheet.
2. Peptides can align in two patterns
a. Parallel orientation- where peptides run in the same direction
(1st peptide strand is CàN and adjacent peptide strand is CàN)
-these are more common
-usually larger structures- consisting many times of more than 5 strands
-distributes hydrophobic side chains on both sides of sheet
b. Anti-Parallel orientation- where peptides run in opposite directions
(1st peptide strand is CàN and adjacent peptide strand is NàC)
-these are less common
-typically smaller structures- as few as 2 strands
-distributes hydrophobic side chains on only one side of the sheet.
c. b-Turn or b-bends
peptide chain forms a sharp bend due to hydrogen bonding of the carbonyl of one residue to the amide
of a residue three positions down the chain.
-typically proline or glycine are found in the b-bend, due to the conformations of the side chain structures.
Image. Alpha helix & beta sheets
Resource (NCBI). 3D structures in proteins.
application: Membrane proteins of known 3D structures
resource: A tour through protein structure
-Defines the conformation (overall shape) the polypeptide takes due to side chain interactions
The conformation that a protein/polypeptide takes is due simply to stabilization. Due to:
1. formation of large number of intra-molecular hydrogen bonds between secondary structures
2. reduction in surface area accessible to a solvent (hydrophobic regions)
a. Disulfide bridges- produced by Cysteine residues
b. Hydrogen bonding- interaction between due to polar or charged side chains
c. Hydrophobic interactions- due to hydrophobic side chains
d. Salt bridges- due to charged side chains
Image: Tertiary bonds
-Three dimensional structure created by interactions of individual peptide sequences (subunits).
a. Types of quaternary structures
1. Dimer- protein consisting 2 polypeptides sequences. Image
2. Trimer, Tetramer, etc.- protein consisting of more than 2 polypeptide sequence
b. Composition
1. homomers- each subunit is identical
2. heteromers- the subunits are different
E. Protein Architecture
There exist 3 major shapes of proteins based on the general conformation and solubility
a. characteristics
1. polypeptide chains are organized parallel along a single axis
2. contain long fibers or large sheets
3. mechanically strong
4. resistant to solubilization in water and dilute salt solutions.
b. examples
a. dominated by a-helices
b. consist of coiled coils-
- a-helix
- coiled coil (2 a-helices)
- protofilament (2 coiled coils)
- filament (4 protofilaments)
c. each a-helix consists of quasi-repeating seven-residue segment (a-b-c-d-e-f-g)
- residues a & d consist of nonpolar residues. These will be oriented to the interior of the helix and
therefore stabilizes the a-helix.
d. some forms of a-helices take advantage of disulfide bridges between helices. Like that in hair (curls)
e. found in claws, fingernails, hair and horns
2. Fibroin- (b-sheets in silk)
- composed of tightly bound b-sheets where each peptide strand contains alternating glycine residues.
This allows other chains to come much closer (those that contain alanine or serine).
3. Collagen – A triple helix
a. basic structural unit is called tropocollagen (contains 3 wound peptides)
- doesn’t form true a-helixes due to the high proline content (30%)
- contains hydroxyprolines (3 & 4)
b. every third residue is Gly- creates the hydrogen bonding between peptides
c. very rigid and inextensible protein
d. found in connective tissue of animals
-generally are spherical in shape and much more numerous
-consist of large amount of a-helices and b-sheets
-pack secondary structures so that only 25% of available volume is space (cavities)
a. examples- almost all enzymes are globular- creation of active site (cavity)
III. Membranous proteins
-membranes that span cellular membrane. Usually have a-helices that span the membrane.
a. Functions of membrane proteins.
Resource: Phosphatidylinositol Signaling
Resources:
Biochemistry from U. Texas. Dr. Rick Russell