| The first step in protein
expression is ligating (inserting) the plasmid into a vector that
contains genes specific for protein expression. The vector Donde
is using is called pTYB2. This has genes which when transformed
into the host will express the protein but also contains genes which
will aid in selecting the appropriate cells which have the modified
phytochrome |
|
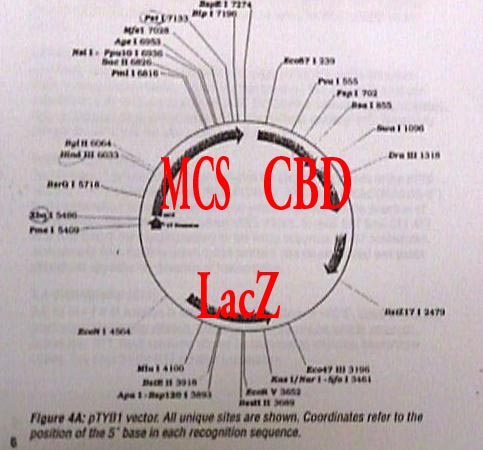
|
The diagram here is of the vector pTYB1 which
is an analog of pTYB2. This vector contains some specialized genes,
seen as dark arc bands, which will help Donde in isolating the mutated phytochrome. |
| MCS |
Multi-Cloning
Site- The position where the
plasmid is inserted into the vector. The lines pointing the vector are
different endonucleases and the sites where each one cuts the vector |
| CBD |
Chitin binding
domain- This site contains a
gene for a protein which will bind with chitin. Chitin (a
polysaccharide) is used in a chromatography column to bind with the
mutated protein complex. |
| LacZ |
b-galactosidase-Reporter
genes are used in vectors to easily identify those cells where the
protein is expressed. LacZ is a gene which synthesizes an enzyme
that breaks down a specific sugar molecule. Cells that do not
possess the recombinant gene produce a blue color whereas in the
nutrient medium, whereas those cells that have expressed the gene
produce the enzyme that breaks down the sugar and is seen as clear or
white. |
|
The faint blue spots are representative
of those cells that do not contain the insert/vector. Some dots on
the plate are white or clear, which means they possess the LacZ gene and
also the mutated plasmid. |
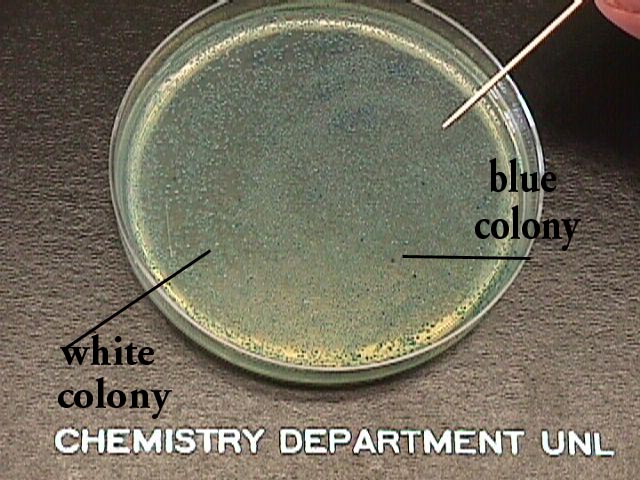
|
|
| The
ligation processes is performed in vitro, but the plasmid (pTYP2 and
insert) next has to be transformed into a host cell, E. coli or P.
pastoris. This involves seeding the cultures, adding different
materials to the nutrient medium which will help in the screening
processes, and finally incubating. See Donde's Subcloning information on
some of these processes. |
|
|
| Once
the cells have been incubated and screened by identifying the cultures
that appear clear, the protein must be isolated. This is done by
lysing the cell using a sonicator (amplified sound waves), centrifuging
the cell components and then filtering using a chitin column. |
|

|

|
| Jeong-Il
is demonstrating the sonicating process to Donde. The sonicator
will break up the cell wall and membranes to produce a mixture of cell
components. This method doesn't use enzymes which could cleave the
mutated phytochrome protein so it is preferred over enzyme digestion. |
|
|
| The columns to
the right contain a polysaccharide, chitin. Chitin is the same
molecule that is found in exoskeletons of insects, spiders and many
crustaceans. This polysaccharide will act as a binding agent for
the new mutated protein complex. The complex contains a
polypeptide segment called Chitin Binding Protein, which is produced
from the Chitin Binding Domain on the vector. This protein will
adhere to the chitin and allow all other materials to wash through the
column, thus selecting only the mutated protein. |

|
|
|
|
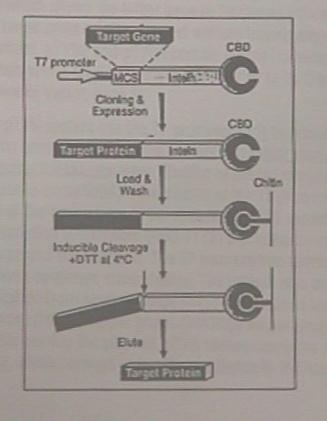
|
This
diagram shows a representation of the mutated protein complex that is
formed from the transformed vector. |
|
The top caption
shows the Vector containing the DNA insert, MCS and CBD. |
|
The vector,
through cloning and expression, will produce the protein complex.
This complex contains the targeted protein (black segment), an intein,
and the chitin binding protein. |
|
The third
caption shows the column chromatography process where chitin is used as
the gel. Note to the right that the chitin binding protein and
chitin are bonded together. This allows all other cell components to be
washed through the gel. |
|
The last step is
to cleave the target protein from the chitin binding protein and intein.
This is done by using a chemical called DTT, Dithiothreitol. This
cleaves the peptide bond between the mutated phytochrome and the intein.
|
|
|
|
| The
final process associated with isolation of the mutated phytochrome is to
verify the protein by SDS-PAGE and Western blot analysis. |
|
|
|
|
|
|





