| Research Procedure | Elizabeth's principle research procedures are to isolate DnaB Helicase, fluorescing single stranded DNA, then titrate the DnaB through florescent ssDNA (single-stranded DNA). | ||
|
|
|||
|
Isolation of DnaB Helicase |
|
Isolation of DnaB helicase starts with genetically altered E.coli which contains a plasmid that overproduces the protein. This sample was obtained from a commercial vendor. | |
|
|
|||
|
|
The first process Elizabeth performed was lysing the cells and then centrifuging to obtain the DnaB helicase. Centrifuging at appropriate speeds allows a researcher to obtain cellular components of known relative masses and other particles can then be discarded. | ||
|
|
|||
|
|
|
||
| Isolation of the DnaB Helicase involves filtering by column gel chromatography. The gel is made of a gelatinous solution that binds molecules according to certain properties (e.g. mass, charge, or molecular affinity). The two gels used here where Heparin-Agarose and Q-Sepharose. The Heparin-Agarose is a gel which acts as a binding medium for proteins that have an affinity for DNA. The Q-Sepharose is a anion exchange medium. This means it will bind those substances which have a net negative charge or negative charge on the surface of the molecule. The gels are then washed with specific solutions having varying concentrations which release the bound protein at different times . | |||
|
|
|||
|
|
|
||
| After the chromatography phase, Elizabeth needed to check for purity of her samples. Gel Electrophoresis (SDS Page) separates the molecules in each sample by properties such as mass and charge. Each band represents a specific molecule or portion of a molecule. Elizabeth is dropping her collected samples from the column chromatography in the apparatus which contains the polyacrylamide gel. These different columns contain different concentrations of the helicase with other proteins. Note the columns to the left of the helicase, the multiple bands refer to other proteins or parts of proteins. | |||
|
|
|||
|
Fluorescing ssDNA |
Elizabeth is using Chloroacetaldehyde to
label single stranded DNA. The labeling compound attaches to the
adenine residues and forms a polyetheno-d-adenine complex. The complex,
when bombarded by light in an luminescence spectrometer, will emit light
at a wavelength about 410 nm.
|
||
|
|
Elizabeth is adding the chloroacetaldehyde to a sample of single stranded DNA which will be placed into the Luminescence Spectrometer. Her results should show an emission around 410 nm. | ||
| To see Elizabeth's write up of the process and the molecules involved, click here. | |||
|
|
|||
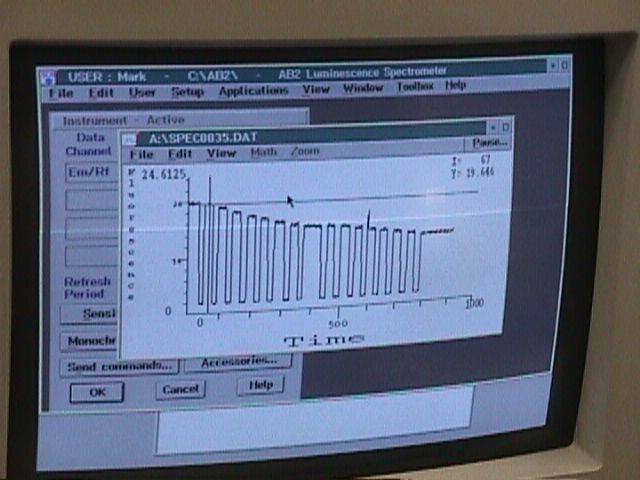
| DnaB Helicase Titration | One important aspect of this research is to identify changes in ssDNA as it binds with the helicase protein. The fluorescent ssDNA emits light of a certain wavelength and when bound to the helicase should produce a conformational change, thus a change in the emission spectra. Dr. Griep believes that the conformation changes arise from a change in nucleotide base stacking. His two hypotheses are that the bases stack tighter (like winding a rope tighter) or loosen up (unwinding a rope) as it binds to the helicase. | ||
|
|
Elizabeth will be using the Fluorometer to titrate varying amount of helicase into her fluorescent ssDNA samples. She will be looking for changes in the fluorescence intensity to identify which of the stacking changes occur. This is a sample titration of fluorescence intensity over time. Notice the decrease in the intensity. Elizabeth will be looking at a sample like this to determine the conformational changes. |
||
|
|
|||